EXPLORATORY ANALYSIS OF LOCAL SPATIAL PATTERNS
Notes from lecture and various articles
Intro
Generally there is very little reason to suppose that a process will be generated randomly over space. Spatial statistics help us to gauge to what extent the values that data take are related to other observations in the vicinity.
Spatial statistics broadly fall into two categories:
1) Global – these allow us to evaluate if there are spatial patterns in the data (clusters)
2) Local – these allow us to evaluate where these spatial patterns are generated
Differences between these two statistics a can be summarized thus:
|
Global |
Local |
| Single Values | Multi-valued |
| Assumed invariant over space | Variant over space |
| Non-mappable | Mappable |
| Used to search for regularities | Used to search for irregularities |
| Aspatial | Spatial |
Generally these statistics are based upon:
- Local means – see spatial weighting sections above (smoothing techniques such as kernel regression and interpolation).
- Covariance methods – comparing the covariances of neighbourhood variables (Moran’s I, and LISA)
- Density methods – the closeness of data points (Ripley’s K, Duranton & Overman’s K-density).
Moran’s I
This is one of the most frequently encountered measures of global association. It is based on the covariance between deviations from the global mean between a data point and its neighbours (howsoever defined – e.g. queen’s/rook’s contiguity at the first/second order etc.).
It is computed in the following way: 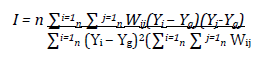
Where there are n data values, y is the outcome variable at location i or its neighbour j, the global mean is Yg and the proximity between locations i and j are given the weights Wij.
A z statistic can be calculated in order to assess the significance of the Moral I estimate (compared in the usual way to a critical value e.g. 1.95 for 5% significance).
Problems with this measure are that it assumes constant variation over space. This may mask a significant amount of heterogeneity in spatial patterns, and it does not allow for local instability of variation. Thus a focus on local patterns of spatial association may be more appropriate. This could involve a decomposition of this type of global indicator in the contribution of each individual observation. One further issue is that the problems associated with MAUP (see above summaries) are built into the Moran statistic.
Local Moran
The Local Moran is a Local Indicator of Spatial Association (LISA) as defined by Anselin (1995). He posits two requirements for a statistic to be considered a LISA:
- The LISA for each observation gives an indication of the extent of spatial clustering of similar values around that observation.
- The sum of the LISAs for all observations is proportional to a global indicator of spatial association.
The local Moran statistic allows us to identify locations where clustering is significant. It may turn out to be similar to the global statistic, but it is equally possible that the local pattern is an aberration in which case the global statistic would not have identified it.
It is calculated like this:
Ii = Zi [∑j=1nWijZj, j=i]
where z are the deviations of observation i or j from the global mean, and w is the weighting system. If I is positive then the location in question has similarly high (low) values as its neighbours, thus forming a cluster.
This statistic can be plotted on the y axis, with the individual observation on the x axis, to investigate outliers, and see whether there is dispersion or clustering.
There are problems with this measure. Firstly the local Moran will be correlated between two locations as the share common elements (neighbours) Due to this problem the usual interpretation of significance will be flawed, hence there is the need for a Bonferroni correction which will correct the significance values (thus reducing the probability of a type I error – wrongly rejecting the null of no clustering). MAUP is an issue similarly as above.
Point Pattern Analysis
This type of analysis looks for patterns in the location of events. This is related to the above techniques, although they are based on aggregated data of which points are the underlying observations. As the analysis is based on disaggregated points, there is no concern about MAUP driving the results.
Ripley’s K
This method counts a firm or other observation’s number of neighbours within a given distance and calculates the average number of neighbours of every firm at every distance – thus a single statistic is calculated for each specified distance. The benchmark test is to look for CSR (complete spatial randomness) which states that observations are located in any place with the same constant probability, and they are so located independently of the location of other observations. This implies a homogenous expected density of points in every part of the territory under examination.
Essentially a circle of given distance (bandwidth) is centred on an observation, and the K statistic is calculated based on all other points that are located within that circle using the following formula:
K(d) = α/n2 * ∑i=n i=1 ∑i≠j I{distanceij < d
where alpha is the area of the study zone (πr2), and I is the count of the points that satisfy the Euclidean distance restriction. If there is an average density of points µ, then the expected number of points in a circle of radius r, is µπr2. As the K statistic is the average number of neighbours divided by the expected number of points µ, this means that CSR leads to K(r) = πr2.
Again, the returned density by distance can be plotted against the uniform distribution to see whether observations are clustered or dispersed relative to CSR.
Marcon and Puech (2003) outline some issues with this measure. Firstly, since the distribution of K is unknown, the variance cannot be evaluated, which necessitates using the Monte Carlo simulation method for constructing confidence intervals. Secondly there are issues at the boundaries of the area studied, as part of the circle will fall outside the boundary (and hence be empty) which may lead to an underestimation at that point. This can be partially corrected for by using only the part of the circle’s area that is under study.
Additionally, CSR is a particularly useful null hypothesis, other benchmarks may be preferable.
Kernel Density
These measures yield local estimates of intensity at a specified point in the study. The most basic form centres a circle on the data point, calculates the number of points in the area and divides by the area of the circle. i.e:
δ(s) = N(C(s, r)/ πr2
where s is the individual observation, N is the number of points within a circle of radius r. The problem with this estimate is that the r is arbitrary, but more seriously, small movements of the circle will cause data points to jump in and out of the estimate which can create discontinuities. One way to improve on this therefore is to specify some weighting scheme where points closer to the centroid contribute more to the calculation than those further away. This type of estimation is called the kernel intensity estimate:
δ(s) = ∑i=n, i=1 1/h2 * k(s – sj / h)
where h is the bandwidth (wider makes estimate more precise, but introduces bias) and K is the kernel weighting function.
